About me
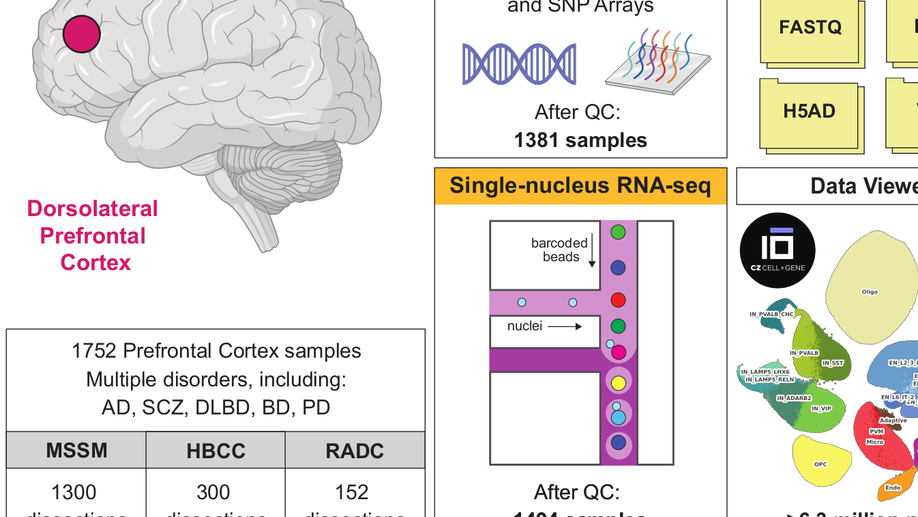
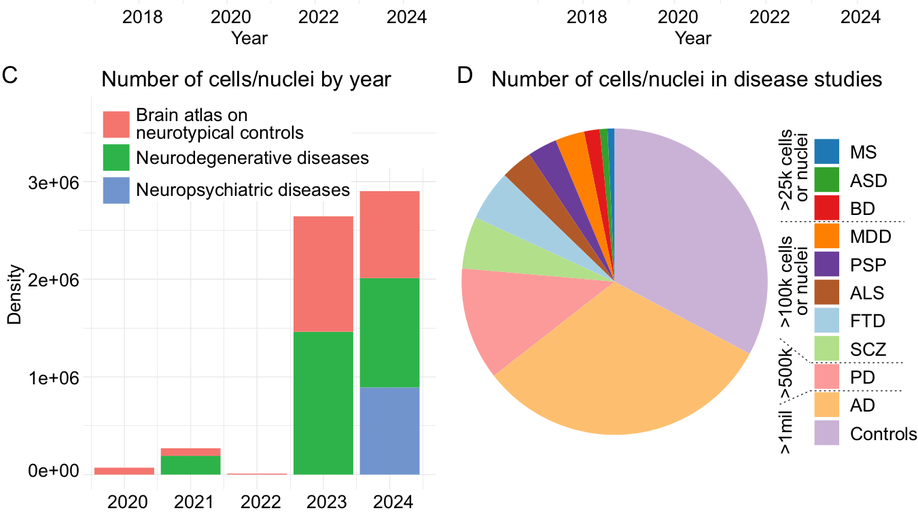
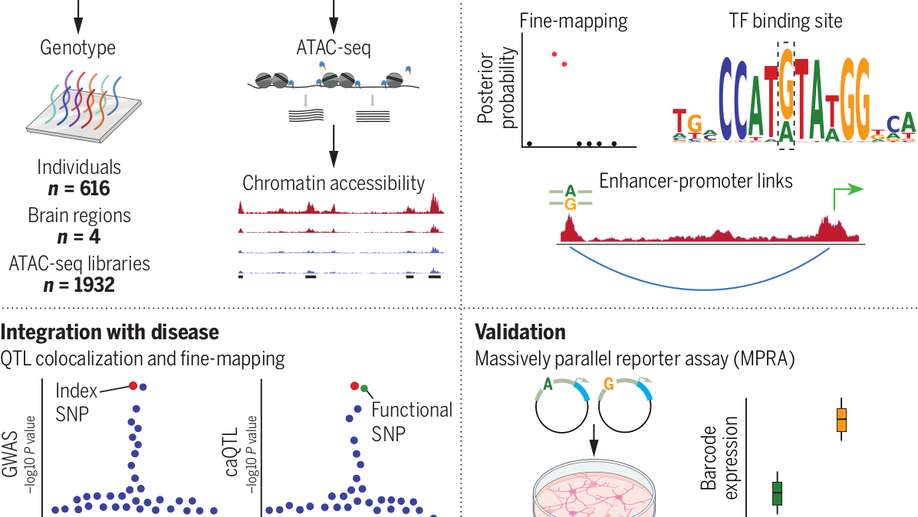
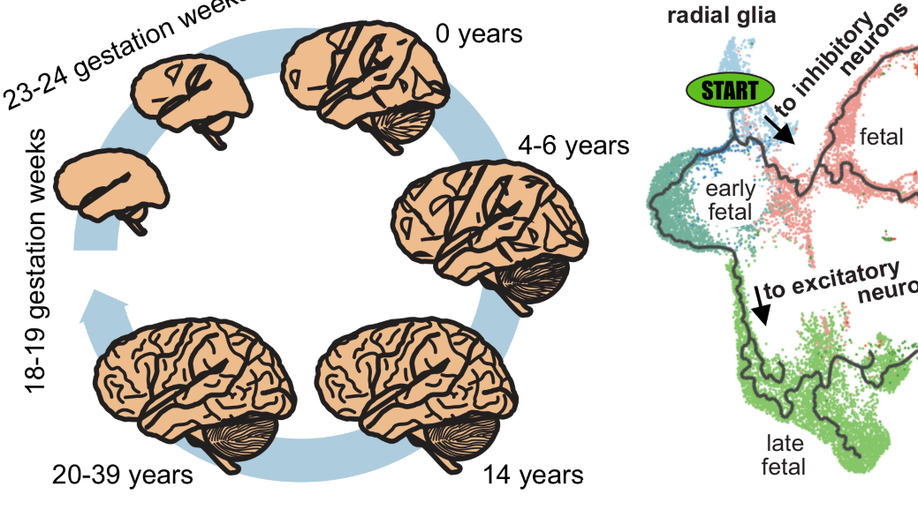
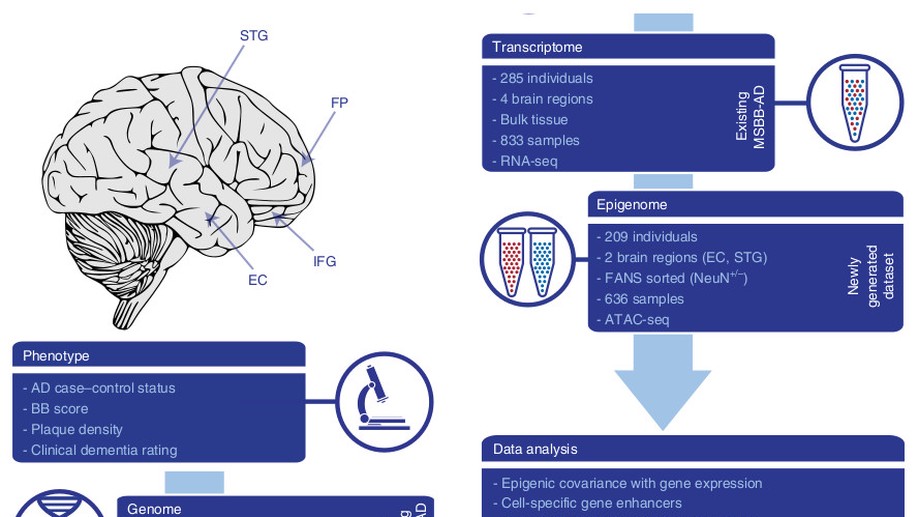
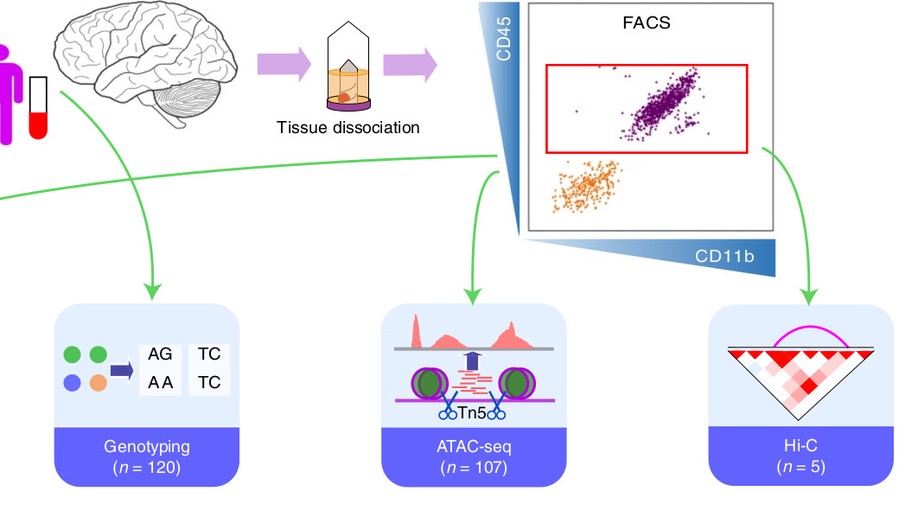
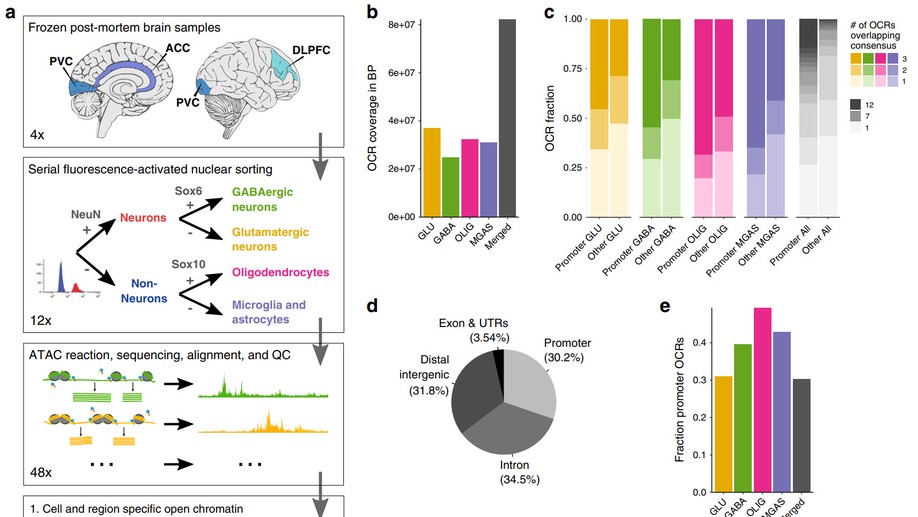
Hey there! My name is Jaroslav Bendl and I am an Assistant Professor at Icahn School of Medicine at Mount Sinai. My long term research interests involve the development of algorithms and pipelines focused on the analysis of the human genome. In my current position, I am specifically focused on neuropsychiatric and neurological disorders such as schizophrenia, depression, bipolar disease, and Alzheimer’s disease. By combining molecular assays generated from postmortem human brains (genome, transcriptome, epigenome), I am trying to come up with a better explanation of genetic factors that carry the risk for those diseases and the mechanisms through which they act.
Interests
- Neuropsychiatric and neurological diseases
- Machine learning
- Rare diseases
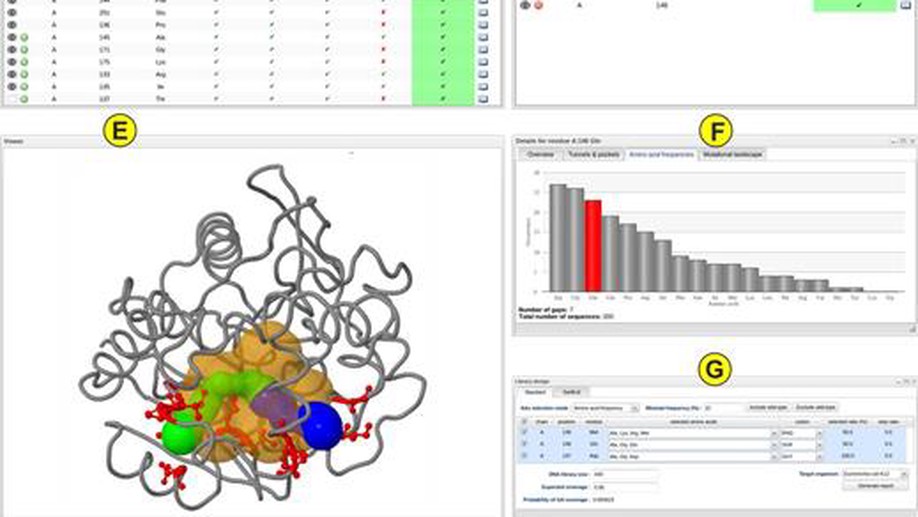
- Protein engineering
- Cellular automata
Education
PhD in Computer Science, 2016
Brno University of Technology & Loschmidt Laboratories, Czechia
MSc in Computer Science, 2011
Brno University of Technology, Czechia
Spring semester internship, 2011
Norwegian University of Science and Technology, Norway
BSc in Computer Science, 2009
Brno University of Technology, Czechia