‘— title: “Population-level variation in enhancer expression identifies disease mechanisms in the human brain”
authors:
- Pengfei Dong
- Gabriel E. Hoffman
- Pasha Apontes
- Jaroslav Bendl
- Samir Rahman
- Michael B. Fernando
- Biao Zeng
- James M. Vicari
- Wen Zhang
- Kiran Girdhar
- Kayla G. Townsley
- Ruth Misir
- the CommonMind Consortium
- Kristen J. Brennand
- Vahram Haroutunian
- Georgios Voloudakis
- John F. Fullard
- Panos Roussos
date: “2022-09-26” doi: “10.1038/s41588-022-01170-4”
Publication type.
Legend: 0 = Uncategorized; 1 = Conference paper; 2 = Journal article;
3 = Preprint / Working Paper; 4 = Report; 5 = Book; 6 = Book section;
7 = Thesis; 8 = Patent
publication_types: [“2”]
Publication name and optional abbreviated publication name.
publication: “Nature Genetics” publication_short: "”
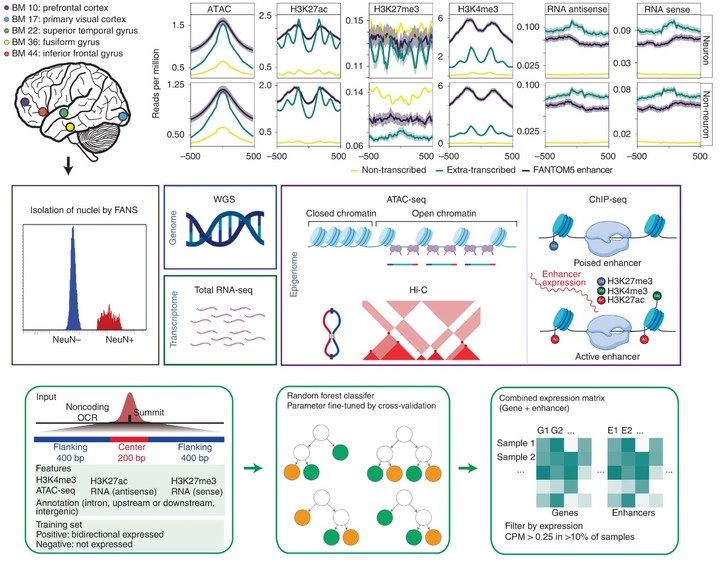
abstract: Identification of risk variants for neuropsychiatric diseases within enhancers underscores the importance of understanding population-level variation in enhancer function in the human brain. Besides regulating tissue-specific and cell-type-specific transcription of target genes, enhancers themselves can be transcribed. By jointly analyzing large-scale cell-type-specific transcriptome and regulome data, we cataloged 30,795 neuronal and 23,265 non-neuronal candidate transcribed enhancers. Examination of the transcriptome in 1,382 brain samples identified robust expression of transcribed enhancers. We explored gene-enhancer coordination and found that enhancer-linked genes are strongly implicated in neuropsychiatric disease. We identified expression quantitative trait loci (eQTLs) for both genes and enhancers and found that enhancer eQTLs mediate a substantial fraction of neuropsychiatric trait heritability. Inclusion of enhancer eQTLs in transcriptome-wide association studies enhanced functional interpretation of disease loci. Overall, our study characterizes the gene-enhancer regulome and genetic mechanisms in the human cortex in both healthy and diseased states.
Summary. An optional shortened abstract.
summary: Analyses of population-level variation in gene and enhancer expression in the human brain characterize the gene–enhancer regulome and the regulatory mechanisms of transcribed enhancers in neuropsychiatric diseases.
tags:
- Source Themes featured: false
links:
- name: "”
url: "”
url_pdf: ‘https://www.nature.com/articles/s41588-022-01170-4' url_code: ‘https://zenodo.org/record/6845955#.Y4OOgdLMJdA' url_dataset: ‘https://doi.org/10.7303/syn25716684' url_poster: '’ url_project: '’ url_slides: '’ url_source: '’ url_video: '’