Abstract
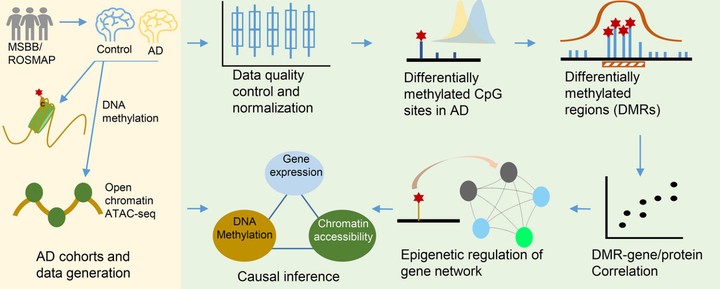
Recent studies revealed the association of abnormal methylomic changes with Alzheimers disease (AD) but there is a lack of systematic study of the impact of methylomic alterations over the molecular networks underlying AD. Methods- We profiled genome-wide methylomic variations in the parahippocampal gyrus from 201 post mortem control, mild cognitive impaired, and AD brains. Results- We identified 270 distinct differentially methylated regions (DMRs) associated with AD. We quantified the impact of these DMRs on each gene and each protein as well as gene and protein co-expression networks. DNA methylation had a profound impact on both AD-associated gene/protein modules and their key regulators. We further integrated the matched multi-omics data to show the impact of DNA methylation on chromatin accessibility, which further modulates gene and protein expression. Discussion- The quantified impact of DNA methylation on gene and protein networks underlying AD identified potential upstream epigenetic regulators of AD.